Introduction
Interleukins (ILs) are a group of cytokines produced by many kinds of cells, which play important roles in transmitting information, activating and regulating immune cells, mediating activation, proliferation and differentiation of T and B cells, and in inflammatory responses. At present, a number of machine learning methods have been proposed to predict ILs inducing peptides, but their predictive performance needs to be further improved, and the inducing peptides of different interleukins are predicted separately, rather than using a general approach.
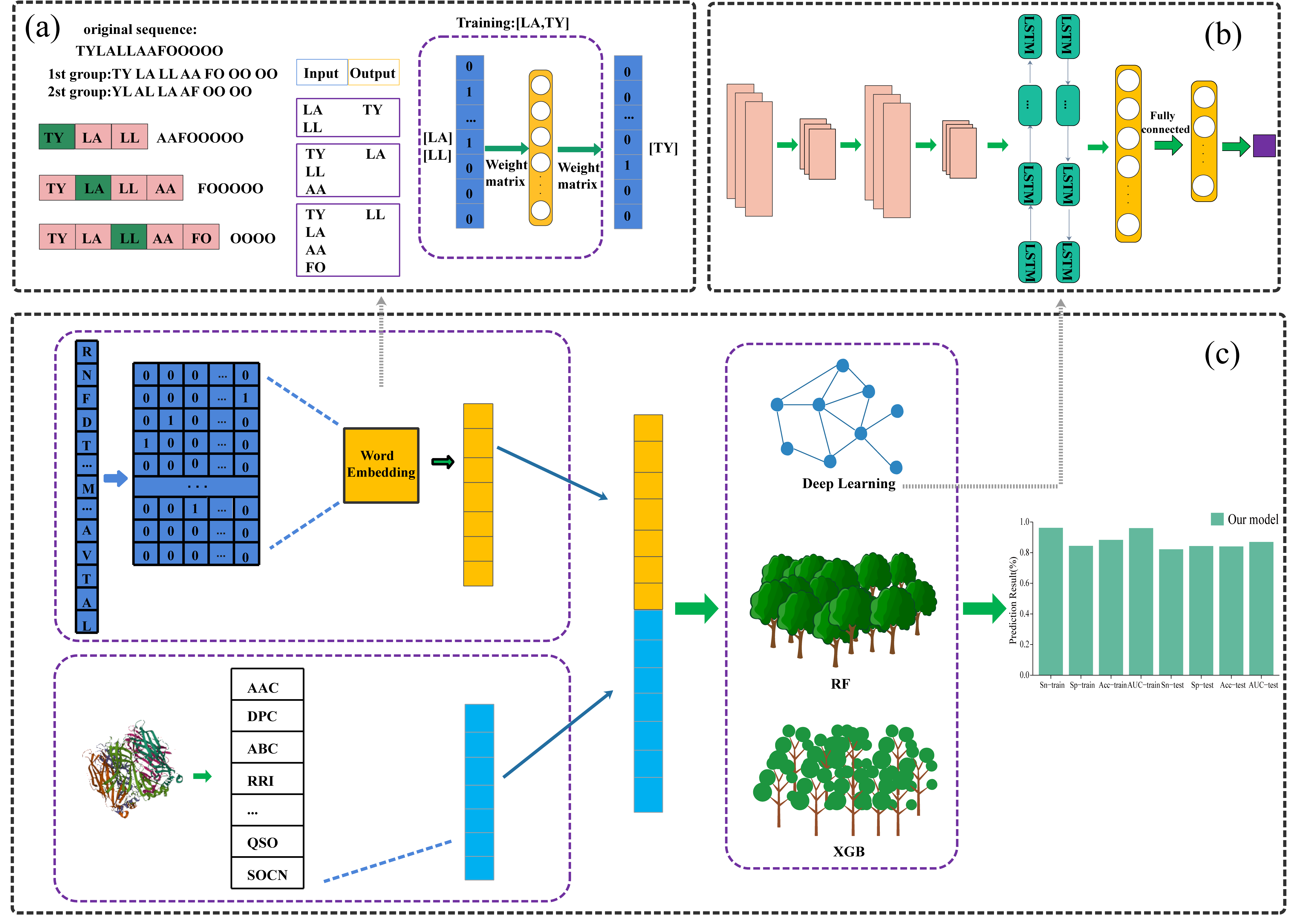
In this work, we combine the statistical features with word embedding of peptide sequence to design an ensemble general model named EnILs to predict inducing peptides of IL-6, IL-10, IL-17, in which the predictive probabilities of random forest, eXtreme Gradient Boosting and neural network are integrated in an average way.
Service
By submitting the data and your contact information, we will return the results to you by email.
Dataset
In this work, the data we used are all extracted from the immune epitope database. For the IL6 Dhall dataset, IL10 Nagpal dataset and IL17 Gupta dataset, sequences containing unnatural amino acids are removed and the lengths of the restriction peptide sequences are 8-25, 8-42 and 5-30, respectively. In addition, we also extracted the unused data of IL6 Dhall dataset, IL10 Nagpal dataset and IL17 Gupta dataset from the immune epitope database as an independent test set, respectively.
The dataset is available for download at the following.
Contact Us
If you have any questions, please feel free to contact us.
Name:
Rui Su
Email:
2909958942@qq.com
Location:
No. 1, Linghai Road, Dalian City, Liaoning Province, China